# !wget https://developer.nvidia.com/compute/cuda/9.0/Prod/local_installers/cuda-repo-ubuntu1604-9-0-local_9.0.176-1_amd64-deb
# !dpkg -i cuda-repo-ubuntu1604-9-0-local_9.0.176-1_amd64-deb
# !apt-key add /var/cuda-repo-9-0-local/7fa2af80.pub
# !apt update -q
# !apt install cuda gcc-6 g++-6 -y -q
# !ln -s /usr/bin/gcc-6 /usr/local/cuda/bin/gcc
# !ln -s /usr/bin/g++-6 /usr/local/cuda/bin/g++
# !curl -sSL "https://julialang-s3.julialang.org/bin/linux/x64/1.7/julia-1.7.3-linux-x86_64.tar.gz" -o julia.tar.gz
# !tar -xzf julia.tar.gz -C /usr --strip-components 1
# !rm -rf julia.tar.gz*
# !julia -e 'using Pkg; pkg"add IJulia; precompile"'
25. Weak IV Experiments#
# Import relevant packages
# Import relevant packages
# using Pkg
# Pkg.add("CSV")
# Pkg.add("DataFrames")
# Pkg.add("GLM")
# Pkg.add("FixedEffectModels")
# Pkg.add("DecisionTree")
# Pkg.add("PrettyTables")
# Pkg.add("CovarianceMatrices")
# Pkg.add("RegressionTables")
# Pkg.add("StatsFuns")
# Pkg.add("Plots")
# Pkg.add("RData")
# Pkg.add("MLBase")
using CSV, DataFrames, FixedEffectModels, DecisionTree, PrettyTables, CovarianceMatrices, RegressionTables, StatsFuns, Plots, RData, MLBase, GLM
function foo1(a;rng=MersenneTwister(3))
return randn(rng,a)
end
foo1 (generic function with 1 method)
function foo2(a;rng=MersenneTwister(1))
return randn(rng,a)
end
foo2 (generic function with 1 method)
B = 1000
IVEst = zeros( B )
n = 100
beta = 0.25
U = foo1(n)
Z = foo2(n)
D = beta*Z+U
Y = D + U;
intercept = ones(length(U))
data1 = DataFrame(intercept = intercept, U = U, Z = Z, D = D, Y = Y);
mod = reg(data1, @formula(D ~ Z))
Linear Model
=======================================================================
Number of obs: 100 Degrees of freedom: 2
R2: 0.067 R2 Adjusted: 0.057
F-Stat: 7.01262 p-value: 0.009
=======================================================================
D | Estimate Std.Error t value Pr(>|t|) Lower 95% Upper 95%
-----------------------------------------------------------------------
Z | 0.250108 0.094447 2.64814 0.009 0.0626815 0.437535
(Intercept) | 0.0873342 0.0949433 0.919856 0.360 -0.101078 0.275746
=======================================================================
IV = reg(data1, @formula(Y ~ 0 + (D ~ Z)))
IV
IV Model
============================================================
Number of obs: 100 Degrees of freedom: 1
R2: 0.752 R2 Adjusted: 0.749
F-Stat: 6.79219 p-value: 0.011
F-Stat (First Stage): 7.01315 p-value (First Stage): 0.008
============================================================
Y | Estimate Std.Error t value Pr(>|t|) Lower 95% Upper 95%
------------------------------------------------------------
D | 0.994509 0.381596 2.60618 0.011 0.23734 1.75168
============================================================
IV.coef
1-element Vector{Float64}:
0.994509337024402
Note that the instrument is weak here (contolled by \(\beta\)) – the t-stat is less than 4.
25.1. Run 1000 trials to evaluate distribution of the IV estimator#
# dependent variable ~ exogenous variables + (endogenous variables ~ instrumental variables)
# Set seed
B = 1000
IVEst = zeros(B)
for i in 1:B
U = randn( n)
Z = randn( n)
D = beta*Z+U
Y = D + U
intercept = ones(length(U))
data2 = DataFrame(intercept = intercept, U = U, Z = Z, D = D, Y = Y);
IV = reg(data2, @formula(Y ~ + (D ~ Z)))
IVEst[i,1] = IV.coef[2]
end
println(minimum(IVEst))
println(maximum(IVEst))
-40.368264648867964
57.83003936193075
IVEst
1000-element Vector{Float64}:
-0.2633141021687235
0.8948832260053269
-0.20787697208091646
0.6792079361265198
1.052671004982911
-0.7421608576444562
1.1668318792400871
0.933747261081341
0.6850752909186767
1.4639180207647025
-0.9642817021998845
-1.497309542643626
1.2189323077695131
⋮
5.460232661324924
0.5478868710622211
1.3717744154929605
1.2644078444676423
1.487865163308266
0.7873831110483491
0.7967961713080116
0.9696312102644875
-0.07785624411680934
0.08917470799234822
1.4232140851370487
0.9943702695396225
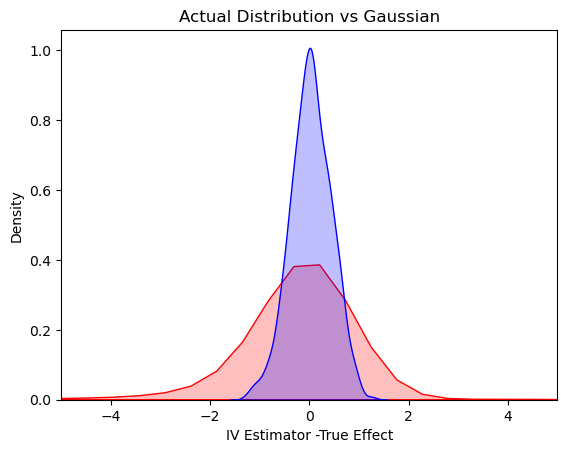
25.2. Plot the Actual Distribution against the Normal Approximation (based on Strong Instrument Assumption)#
val = collect(range( -5, 5.5, step = 0.05 ))
var = (1/beta^2)*(1/100) # theoretical variance of IV
sd = sqrt(var)
μ=0; σ=sd
d = Normal(μ, σ)
normal_dist = rand(d,1000)
# plotting both distibutions on the same figure
Seaborn.kdeplot(x = IVEst.-1, shade = true, color = "red")
Seaborn.kdeplot(x = normal_dist, shade = true, color = "blue")
Seaborn.title("Actual Distribution vs Gaussian")
Seaborn.xlabel("IV Estimator -True Effect")
Seaborn.xlim(-5,5)
(-5.0, 5.0)

