23. Sensitivity Analysis with Sensmakr and Debiased ML#
23.1. Sensititivy Analysis for Unobserved Confounder with DML and Sensmakr#
Here we experiment with using package “sensemakr” in conjunction with debiased ML
We will
mimic the partialling out procedure with machine learning tools,
and invoke Sensmakr to compute \(\phi^2\) and plot sensitivity results.
We will use the sensemakr package adapted in python (PySensemakr) by Brian Hill and Nathan LaPierre ink
#pip install PySensemakr
#Import packages
import sensemakr as smkr
import statsmodels.api as sm
import statsmodels.formula.api as smf
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from sklearn.model_selection import KFold
import warnings
warnings.filterwarnings('ignore')
darfur = pd.read_csv("https://raw.githubusercontent.com/d2cml-ai/14.388_py/main/data/darfur.csv")
darfur.shape
(1276, 14)
Data is described here https://cran.r-project.org/web/packages/sensemakr/vignettes/sensemakr.html
The main outcome is attitude towards peace – the peacefactor. The key variable of interest is whether the responders were directly harmed (directlyharmed). We want to know if being directly harmed in the conflict causes people to support peace-enforcing measures. The measured confounders include female indicator, age, farmer, herder, voted in the past, and household size. There is also a village indicator, which we will treat as fixed effect and partial it out before conducting the analysis. The standard errors will be clustered at the village level.
23.2. Take out village fixed effects and run basic linear analysis#
# get rid of village fixed effects
import statsmodels.api as sm
import statsmodels.formula.api as smf
# 1. basic model
peacefactorR = smf.ols('peacefactor~village' , data=darfur).fit().resid
directlyharmedR = smf.ols('directlyharmed~village' , data=darfur).fit().resid
femaleR = smf.ols('female~village' , data=darfur).fit().resid
ageR = smf.ols('age~village' , data=darfur).fit().resid
farmerR = smf.ols('farmer_dar~village' , data=darfur).fit().resid
herderR = smf.ols('herder_dar~village' , data=darfur).fit().resid
pastvotedR = smf.ols('pastvoted~village' , data=darfur).fit().resid
hhsizeR = smf.ols('hhsize_darfur~village' , data=darfur).fit().resid
darfurR = pd.concat([peacefactorR, directlyharmedR, femaleR,
ageR, farmerR, herderR, pastvotedR,
hhsizeR, darfur['village']], axis=1)
darfurR.head()
| 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | village | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | 1.310063e-14 | 5.171073e-14 | -1.353145e-13 | 2.749800e-12 | -8.823583e-14 | -7.293814e-15 | -6.172840e-14 | 1.431744e-12 | Abdel Khair |
| 1 | -9.903189e-14 | 7.581381e-14 | 7.049916e-14 | 9.912071e-13 | 1.043610e-13 | 2.568140e-14 | -4.152234e-14 | -1.957545e-12 | Abdi Dar |
| 2 | -3.029464e-01 | -3.333333e-01 | -3.333333e-01 | 6.666667e-01 | 3.333333e-01 | 6.646498e-17 | -6.666667e-01 | 4.666667e+00 | Abu Sorog |
| 3 | 3.301185e-01 | 3.333333e-01 | -6.666667e-01 | 1.733333e+01 | -6.666667e-01 | -3.333333e-01 | -3.333333e-01 | 1.666667e+00 | Abu Dejaj |
| 4 | -1.650592e-01 | 3.333333e-01 | 3.333333e-01 | -1.266667e+01 | 3.333333e-01 | -3.333333e-01 | 6.666667e-01 | -3.333333e-01 | Abu Dejaj |
darfurR.columns = ["peacefactorR", "directlyharmedR", "femaleR",
"ageR", "farmerR", "herderR", "pastvotedR",
"hhsize_darfurR", "village"]
darfurR.head()
| peacefactorR | directlyharmedR | femaleR | ageR | farmerR | herderR | pastvotedR | hhsize_darfurR | village | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | 1.310063e-14 | 5.171073e-14 | -1.353145e-13 | 2.749800e-12 | -8.823583e-14 | -7.293814e-15 | -6.172840e-14 | 1.431744e-12 | Abdel Khair |
| 1 | -9.903189e-14 | 7.581381e-14 | 7.049916e-14 | 9.912071e-13 | 1.043610e-13 | 2.568140e-14 | -4.152234e-14 | -1.957545e-12 | Abdi Dar |
| 2 | -3.029464e-01 | -3.333333e-01 | -3.333333e-01 | 6.666667e-01 | 3.333333e-01 | 6.646498e-17 | -6.666667e-01 | 4.666667e+00 | Abu Sorog |
| 3 | 3.301185e-01 | 3.333333e-01 | -6.666667e-01 | 1.733333e+01 | -6.666667e-01 | -3.333333e-01 | -3.333333e-01 | 1.666667e+00 | Abu Dejaj |
| 4 | -1.650592e-01 | 3.333333e-01 | 3.333333e-01 | -1.266667e+01 | 3.333333e-01 | -3.333333e-01 | 6.666667e-01 | -3.333333e-01 | Abu Dejaj |
# Preliminary linear model analysis
# Linear model 1
linear_model_1 = smf.ols('peacefactorR~ directlyharmedR+ femaleR + ageR + farmerR+ herderR + pastvotedR + hhsizeR'
,data=darfurR ).fit().get_robustcov_results(cov_type = "cluster", groups= darfurR['village'])
linear_model_1_table = linear_model_1.summary2().tables[1]
linear_model_1_table
| Coef. | Std.Err. | t | P>|t| | [0.025 | 0.975] | |
|---|---|---|---|---|---|---|
| Intercept | -2.538768e-15 | 2.685827e-15 | -0.945246 | 3.450038e-01 | -7.816060e-15 | 2.738525e-15 |
| directlyharmedR | 9.731582e-02 | 2.382281e-02 | 4.084985 | 5.156752e-05 | 5.050716e-02 | 1.441245e-01 |
| femaleR | -2.320514e-01 | 2.443857e-02 | -9.495295 | 1.000143e-19 | -2.800700e-01 | -1.840329e-01 |
| ageR | -2.071749e-03 | 7.441260e-04 | -2.784137 | 5.576761e-03 | -3.533858e-03 | -6.096402e-04 |
| farmerR | -4.044295e-02 | 2.956411e-02 | -1.367974 | 1.719536e-01 | -9.853250e-02 | 1.764661e-02 |
| herderR | 1.427910e-02 | 3.649802e-02 | 0.391229 | 6.957994e-01 | -5.743466e-02 | 8.599286e-02 |
| pastvotedR | -4.802496e-02 | 2.687661e-02 | -1.786868 | 7.458324e-02 | -1.008339e-01 | 4.784016e-03 |
| hhsizeR | 1.229812e-03 | 2.166312e-03 | 0.567698 | 5.705024e-01 | -3.026704e-03 | 5.486328e-03 |
# Linear model 2
linear_model_2 = smf.ols('peacefactorR~ femaleR + ageR + farmerR+ herderR + pastvotedR + hhsizeR'
,data=darfurR ).fit().get_robustcov_results(cov_type = "cluster", groups= darfurR['village'])
linear_model_2_table = linear_model_2.summary2().tables[1]
linear_model_2_table
| Coef. | Std.Err. | t | P>|t| | [0.025 | 0.975] | |
|---|---|---|---|---|---|---|
| Intercept | -2.465909e-15 | 2.453072e-15 | -1.005233 | 3.152858e-01 | -7.285870e-15 | 2.354052e-15 |
| femaleR | -2.415042e-01 | 2.536306e-02 | -9.521887 | 8.063158e-20 | -2.913393e-01 | -1.916692e-01 |
| ageR | -2.186810e-03 | 7.453429e-04 | -2.933966 | 3.505173e-03 | -3.651310e-03 | -7.223106e-04 |
| farmerR | -4.071431e-02 | 2.923021e-02 | -1.392885 | 1.642928e-01 | -9.814780e-02 | 1.671917e-02 |
| herderR | 2.622875e-02 | 3.967810e-02 | 0.661038 | 5.089015e-01 | -5.173345e-02 | 1.041909e-01 |
| pastvotedR | -4.414131e-02 | 2.784297e-02 | -1.585367 | 1.135348e-01 | -9.884904e-02 | 1.056643e-02 |
| hhsizeR | 1.336220e-03 | 2.126678e-03 | 0.628314 | 5.300943e-01 | -2.842419e-03 | 5.514859e-03 |
# Linear model 3
linear_model_3 = smf.ols('directlyharmedR~ femaleR + ageR + farmerR+ herderR + pastvotedR + hhsizeR'
,data=darfurR ).fit().get_robustcov_results(cov_type = "cluster", groups= darfurR['village'])
linear_model_3_table = linear_model_3.summary2().tables[1]
linear_model_3_table
| Coef. | Std.Err. | t | P>|t| | [0.025 | 0.975] | |
|---|---|---|---|---|---|---|
| Intercept | 8.380883e-16 | 2.459246e-15 | 0.340791 | 0.733409 | -3.994003e-15 | 5.670179e-15 |
| femaleR | -9.713517e-02 | 5.128637e-02 | -1.893976 | 0.058823 | -1.979061e-01 | 3.635740e-03 |
| ageR | -1.182350e-03 | 1.150680e-03 | -1.027523 | 0.304687 | -3.443283e-03 | 1.078584e-03 |
| farmerR | -2.788538e-03 | 4.280159e-02 | -0.065150 | 0.948081 | -8.688797e-02 | 8.131090e-02 |
| herderR | 1.227925e-01 | 5.064352e-02 | 2.424643 | 0.015688 | 2.328466e-02 | 2.223002e-01 |
| pastvotedR | 3.990773e-02 | 3.366253e-02 | 1.185524 | 0.236391 | -2.623468e-02 | 1.060501e-01 |
| hhsizeR | 1.093431e-03 | 3.286160e-03 | 0.332738 | 0.739476 | -5.363437e-03 | 7.550298e-03 |
23.3. We first use Lasso for Partilling Out Controls#
import hdmpy
import patsy
from patsy import ModelDesc, Term, EvalFactor
X = patsy.dmatrix("(femaleR + ageR + farmerR+ herderR + pastvotedR + hhsizeR)**3", darfurR)
Y = darfurR['peacefactorR'].to_numpy()
D = darfurR['directlyharmedR'].to_numpy()
resY = hdmpy.rlasso(X,Y, post = False).est['residuals'].reshape( Y.size,)
resD = hdmpy.rlasso(X,D, post = False).est['residuals'].reshape( D.size,)
FVU_Y = 1 - np.var(resY)/np.var(peacefactorR)
FVU_D = 1 - np.var(resD)/np.var(directlyharmedR)
print("Controls explain the following fraction of variance of Outcome", FVU_Y)
print("Controls explain the following fraction of variance of treatment", FVU_D)
Controls explain the following fraction of variance of Outcome 0.12505767421787006
Controls explain the following fraction of variance of treatment 0.011939558050506838
darfurR['resY'] = resY
darfurR['resD'] = resD
# Filan estimation
# Culster SE by village
dml_darfur_model = smf.ols('resY~ resD',data=darfurR ).fit().get_robustcov_results(cov_type = "cluster", groups= darfurR['village'])
dml_darfur_model_table = dml_darfur_model.summary2().tables[1]
dml_darfur_model_table
| Coef. | Std.Err. | t | P>|t| | [0.025 | 0.975] | |
|---|---|---|---|---|---|---|
| Intercept | -6.357952e-10 | 0.000159 | -0.000004 | 0.999997 | -0.000312 | 0.000312 |
| resD | 1.003438e-01 | 0.024429 | 4.107623 | 0.000047 | 0.052345 | 0.148343 |
23.4. Manual Bias Analysis#
# linear model to use as input in sensemakr
dml_darfur_model= smf.ols('resY~ resD',data=darfurR ).fit()
dml_darfur_model_table = dml_darfur_model.summary2().tables[1]
dml_darfur_model_table
| Coef. | Std.Err. | t | P>|t| | [0.025 | 0.975] | |
|---|---|---|---|---|---|---|
| Intercept | -6.357952e-10 | 0.006843 | -9.291193e-08 | 9.999999e-01 | -0.013425 | 0.013425 |
| resD | 1.003438e-01 | 0.018273 | 5.491364e+00 | 4.809577e-08 | 0.064495 | 0.136192 |
beta = dml_darfur_model_table['Coef.'][1]
beta
0.10034379376801948
# Hypothetical values of partial R2s
R2_YC = .16
R2_DC = .01
# Elements of the formal
kappa = (R2_YC * R2_DC)/(1- R2_DC)
varianceRatio = np.mean(dml_darfur_model.resid**2)/np.mean(dml_darfur_model.resid**2)
# Compute square bias
BiasSq = kappa*varianceRatio
# Compute absolute value of the bias
print(np.sqrt(BiasSq))
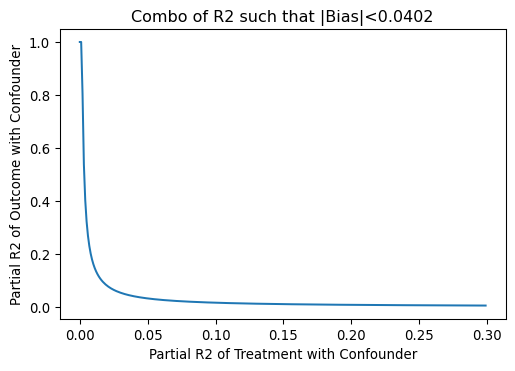
# plotting
gridR2_DC = np.arange(0,0.3,0.001)
gridR2_YC = kappa*(1 - gridR2_DC)/gridR2_DC
gridR2_YC = np.where(gridR2_YC > 1, 1, gridR2_YC)
plt.title("Combo of R2 such that |Bias|<{}".format(round(np.sqrt(BiasSq), 5)))
plt.xlabel("Partial R2 of Treatment with Confounder")
plt.ylabel("Partial R2 of Outcome with Confounder")
plt.plot(gridR2_DC,gridR2_YC)
plt.show()
0.04020151261036848
23.5. Bias Analysis with Sensemakr#
# Imports
import sensemakr as smkr
# from sensemakr import sensitivity_stats
# from sensemakr import bias_functions
# from sensemakr import ovb_bounds
# from sensemakr import ovb_plots
import statsmodels.api as sm
import statsmodels.formula.api as smf
import numpy as np
import pandas as pd
a = 1
b = 3
if a is not None and b is not None:
print('hola')
hola
import sensemakr as smkr
# We need to double check why the function does not allow to run withour the benchmark_covariates argument
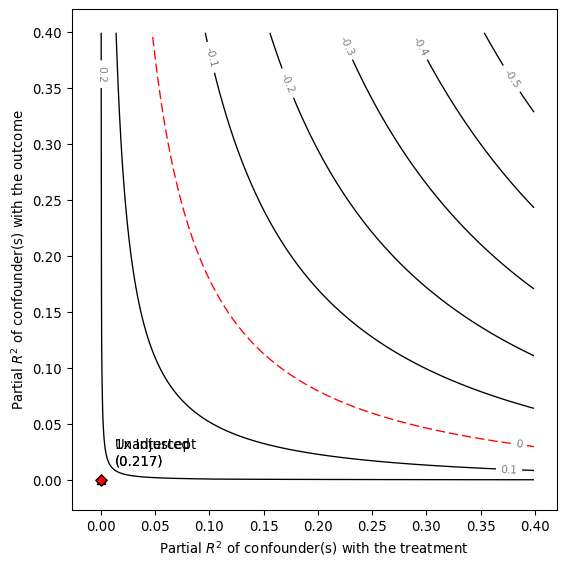
model = smkr.Sensemakr( model = dml_darfur_model, treatment = "resD")
model.summary()
Sensitivity Analysis to Unobserved Confounding
Model Formula: resY~ resD
Null hypothesis: q = 1 and reduce = True
-- This means we are considering biases that reduce the absolute value of the current estimate.
-- The null hypothesis deemed problematic is H0:tau = 0.0
Unadjusted Estimates of ' resD ':
Coef. estimate: 0.1
Standard Error: 0.018
t-value: 5.491
Sensitivity Statistics:
Partial R2 of treatment with outcome: 0.023
Robustness Value, q = 1 : 0.142
Robustness Value, q = 1 alpha = 0.05 : 0.094
Verbal interpretation of sensitivity statistics:
-- Partial R2 of the treatment with the outcome: an extreme confounder (orthogonal to the covariates) that explains 100% of the residual variance of the outcome, would need to explain at least 2.312 % of the residual variance of the treatment to fully account for the observed estimated effect.
-- Robustness Value, q = 1 : unobserved confounders (orthogonal to the covariates) that explain more than 14.247 % of the residual variance of both the treatment and the outcome are strong enough to bring the point estimate to 0.0 (a bias of 100.0 % of the original estimate). Conversely, unobserved confounders that do not explain more than 14.247 % of the residual variance of both the treatment and the outcome are not strong enough to bring the point estimate to 0.0 .
-- Robustness Value, q = 1 , alpha = 0.05 : unobserved confounders (orthogonal to the covariates) that explain more than 9.41 % of the residual variance of both the treatment and the outcome are strong enough to bring the estimate to a range where it is no longer 'statistically different' from 0.0 (a bias of 100.0 % of the original estimate), at the significance level of alpha = 0.05 . Conversely, unobserved confounders that do not explain more than 9.41 % of the residual variance of both the treatment and the outcome are not strong enough to bring the estimate to a range where it is no longer 'statistically different' from 0.0 , at the significance level of alpha = 0.05 .
model.plot()
23.6. Next We use Random Forest as ML tool for Partialling Out#
The following code does DML with clsutered standard errors by ClusterID
import itertools
from itertools import compress
def DML2_for_PLM(x, d, y, dreg, yreg, nfold, clu):
kf = KFold(n_splits = nfold, shuffle=True) #Here we use kfold to generate kfolds
I = np.arange(0, len(d)) #To have a id vector from data
train_id, test_id = [], [] #arrays to store kfold's ids
#generate and store kfold's id
for kfold_index in kf.split(I):
train_id.append(kfold_index[0])
test_id.append(kfold_index[1])
# Create array to save errors
dtil = np.zeros( len(y) ).reshape( len(d) , 1 )
ytil = np.zeros( len(y) ).reshape( len(d) , 1 )
# loop to save results
for b in range(0, len(train_id)):
# Lasso regression, excluding folds selected
dfit = dreg(x[train_id[b],], d[train_id[b],])
yfit = yreg(x[train_id[b],], y[train_id[b],])
# predict estimates using the
dhat = dfit.predict( x[test_id[b],] )
yhat = yfit.predict( x[test_id[b],] )
# save errors
dtil[test_id[b]] = d[test_id[b],] - dhat.reshape( -1 , 1 )
ytil[test_id[b]] = y[test_id[b],] - yhat.reshape( -1 , 1 )
print(b, " ")
# Create dataframe
data_2 = pd.DataFrame(np.concatenate( ( ytil, dtil,clu ), axis = 1), columns = ['ytil','dtil','CountyCode'])
# OLS clustering at the County level
model = "ytil ~ dtil"
baseline_ols = smf.ols(model , data=data_2).fit().get_robustcov_results(cov_type = "cluster", groups= data_2['CountyCode'])
coef_est = baseline_ols.summary2().tables[1]['Coef.']['dtil']
se = baseline_ols.summary2().tables[1]['Std.Err.']['dtil']
print("Coefficient is {}, SE is equal to {}".format(coef_est, se))
return coef_est, se, dtil, ytil, data_2
#return dtil, ytil, data_2
from sklearn.tree import DecisionTreeRegressor
from sklearn.ensemble import RandomForestRegressor
from sklearn.ensemble import GradientBoostingRegressor
from sklearn.preprocessing import LabelEncoder
# This new matrix include intercept
x = patsy.dmatrix("~ femaleR + ageR + farmerR + herderR + pastvotedR + hhsizeR", darfurR)
y = darfurR['peacefactorR'].to_numpy().reshape( len(Y) , 1 )
d = darfurR['directlyharmedR'].to_numpy().reshape( len(Y) , 1 )
darfurR['village'].unique().size
486
# creating instance of labelencoder
labelencoder = LabelEncoder()
# Assigning numerical values and storing in another column
darfurR['village_clu'] = labelencoder.fit_transform(darfurR['village'])
# Create cluster object
CLU = darfurR['village_clu']
clu = CLU.to_numpy().reshape( len(Y) , 1 )
#DML with RF
def dreg(x,d):
result = RandomForestRegressor( random_state = 0,
n_estimators = 500,
max_features = max( int( x.shape[1] / 3 ), 1 ),
min_samples_leaf = 1 ).fit( x, d )
return result
def yreg(x,y):
result = RandomForestRegressor( random_state = 0,
n_estimators = 500,
max_features = max( int( x.shape[1] / 3 ), 1 ),
min_samples_leaf = 1 ).fit( x, y )
return result
DML2_RF = DML2_for_PLM(x, d, y, dreg, yreg, 10, clu) # set to 2 due to computation time
0
1
2
3
4
5
6
7
8
9
Coefficient is 0.09012697712229308, SE is equal to 0.02391945640215873
resY = DML2_RF[2]
resD = DML2_RF[3]
FVU_Y = max(1 - ( np.var(resY)/np.var(peacefactorR) ), 0 )
FVU_D = max(1 - ( np.var(resD)/np.var(directlyharmedR) ), 0 )
print("Controls explain the following fraction of variance of Outcome", FVU_Y)
print("Controls explain the following fraction of variance of treatment", FVU_D)
Controls explain the following fraction of variance of Outcome 0
Controls explain the following fraction of variance of treatment 0.5258708374482872
darfurR['resY_rf'] = resY
darfurR['resD_rf'] = resD
# linear model to use as input in sensemakr
dml_darfur_model_rf= smf.ols('resY_rf~ resD_rf',data=darfurR ).fit()
dml_darfur_model_rf_table = dml_darfur_model_rf.summary2().tables[1]
# We need to double check why the function does not allow to run withour the benchmark_covariates argument
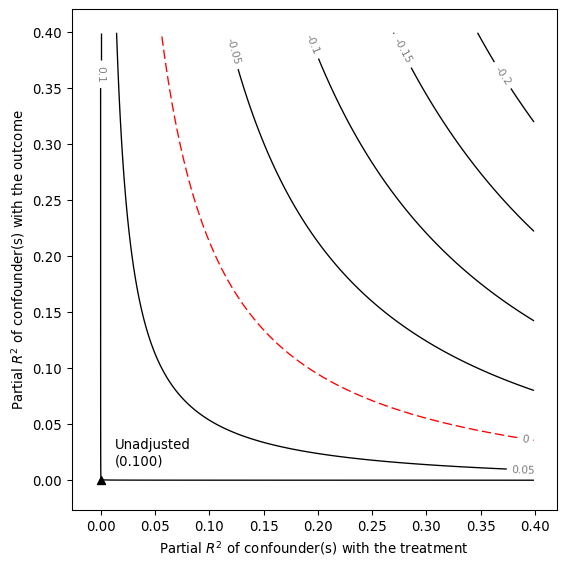
dml_darfur_sensitivity = smkr.Sensemakr(dml_darfur_model_rf, "resD_rf", benchmark_covariates = "Intercept")
dml_darfur_sensitivity.summary()
# Make a contour plot for the estimate
dml_darfur_sensitivity.plot()
Sensitivity Analysis to Unobserved Confounding
Model Formula: resY_rf~ resD_rf
Null hypothesis: q = 1 and reduce = True
-- This means we are considering biases that reduce the absolute value of the current estimate.
-- The null hypothesis deemed problematic is H0:tau = 0.0
Unadjusted Estimates of ' resD_rf ':
Coef. estimate: 0.217
Standard Error: 0.043
t-value: 5.039
Sensitivity Statistics:
Partial R2 of treatment with outcome: 0.02
Robustness Value, q = 1 : 0.132
Robustness Value, q = 1 alpha = 0.05 : 0.083
Verbal interpretation of sensitivity statistics:
-- Partial R2 of the treatment with the outcome: an extreme confounder (orthogonal to the covariates) that explains 100% of the residual variance of the outcome, would need to explain at least 1.954 % of the residual variance of the treatment to fully account for the observed estimated effect.
-- Robustness Value, q = 1 : unobserved confounders (orthogonal to the covariates) that explain more than 13.155 % of the residual variance of both the treatment and the outcome are strong enough to bring the point estimate to 0.0 (a bias of 100.0 % of the original estimate). Conversely, unobserved confounders that do not explain more than 13.155 % of the residual variance of both the treatment and the outcome are not strong enough to bring the point estimate to 0.0 .
-- Robustness Value, q = 1 , alpha = 0.05 : unobserved confounders (orthogonal to the covariates) that explain more than 8.254 % of the residual variance of both the treatment and the outcome are strong enough to bring the estimate to a range where it is no longer 'statistically different' from 0.0 (a bias of 100.0 % of the original estimate), at the significance level of alpha = 0.05 . Conversely, unobserved confounders that do not explain more than 8.254 % of the residual variance of both the treatment and the outcome are not strong enough to bring the estimate to a range where it is no longer 'statistically different' from 0.0 , at the significance level of alpha = 0.05 .
Bounds on omitted variable bias:
--The table below shows the maximum strength of unobserved confounders with association with the treatment and the outcome bounded by a multiple of the observed explanatory power of the chosen benchmark covariate(s).
bound_label r2dz_x r2yz_dx treatment adjusted_estimate adjusted_se \
0 1x Intercept 0.000105 0.000035 resD_rf 0.216687 0.043043
adjusted_t adjusted_lower_CI adjusted_upper_CI
0 5.034236 0.132245 0.301129