9. Experiment on orthogonal Learning#
9.1. Simulation Design 2#
# install.packages("librarian", quiet = T)
librarian::shelf(hdm, quiet = T)
set.seed(1)
B= 100 # trials
Naive = rep(0, B)
Orthogonal = rep(0, B)
for (i in 1:B){
n = 10000
p = 100
beta = 1 / (1:p)^2
gamma =1 / (1:p)^2
X = matrix(rnorm(n*p), n, p)
D = X%*%gamma + rnorm(n)/4
Y = D + X%*%beta + rnorm(n)
# single selection method
SX.IDs = which(rlasso(Y~ D+X)$coef[-c(1,2)] !=0) #select covariates by Lasso
if (sum(SX.IDs)==0) {Naive[i] = lm(Y~ D)$coef[2]}
if (sum(SX.IDs)>0) {Naive[i] = lm(Y~ D + X[,SX.IDs])$coef[2]}
#partialling out
resY = rlasso(Y~ X, Post=F)$res
resD = rlasso(D~ X, Post=F)$res
Orthogonal[i]= lm(resY ~ resD)$coef[2]
}
Installing package into ‘/usr/local/lib/R/site-library’
(as ‘lib’ is unspecified)
also installing the dependency ‘BiocManager’
These packages will be installed:
'hdm'
It may take some time.
also installing the dependencies ‘iterators’, ‘foreach’, ‘shape’, ‘Rcpp’, ‘RcppEigen’, ‘glmnet’, ‘checkmate’, ‘Formula’
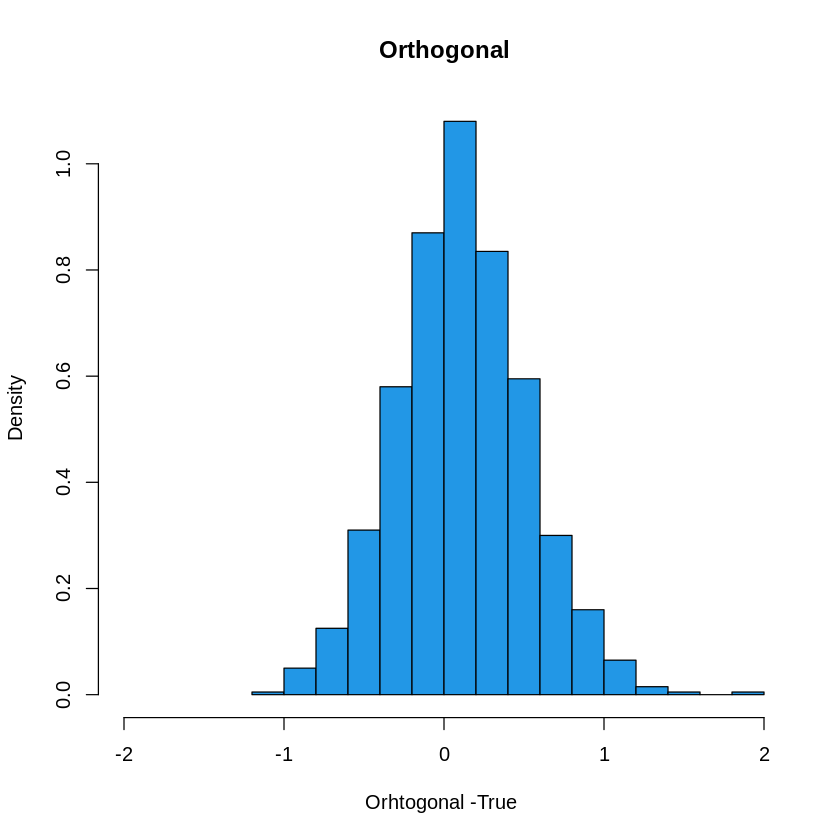
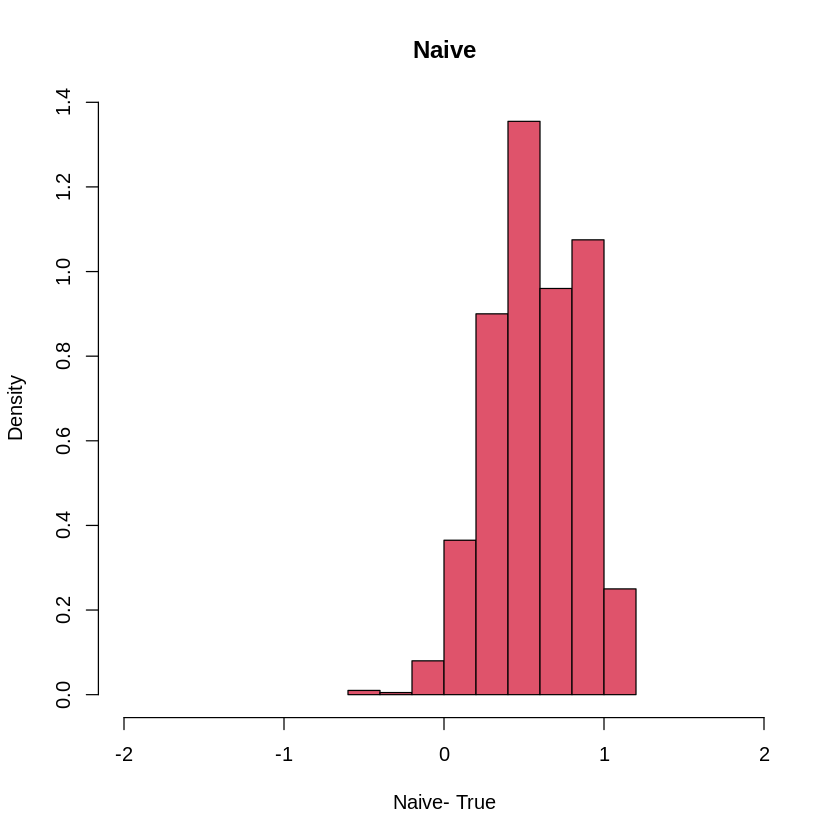
hist(Orthogonal-1,col=4, freq=F, xlim= c(-2, 2), xlab= "Orhtogonal -True ", main="Orthogonal")
hist(Naive-1, col=2, freq=F, xlim= c(-2,2), xlab= "Naive- True", main = "Naive")
9.2. Simulation Design 2#
# library(hdm)
set.seed(1)
B= 1000 # trials
Naive = rep(0, B)
Orthogonal = rep(0, B)
for (i in 1:B){
n =100
p = 100
beta = 1/(1:p)^2
gamma = 1/(1:p)^2
X = matrix(rnorm(n*p), n, p)
D = X%*%gamma + rnorm(n)/4
Y = D+ X%*%beta + rnorm(n)
# single selection method
SX.IDs = which(rlasso(Y~ D+X)$coef[-c(1,2)] !=0) #select covariates by Lasso
if (sum(SX.IDs)==0) {Naive[i] = lm(Y~ D)$coef[2]}
if (sum(SX.IDs)>0) {Naive[i] = lm(Y~ D + X[,SX.IDs])$coef[2]}
#partialling out
resY = rlasso(Y~ X, Post=T)$res
resD = rlasso(D~ X, Post=T)$res
Orthogonal[i]= lm(resY ~ resD)$coef[2]
}
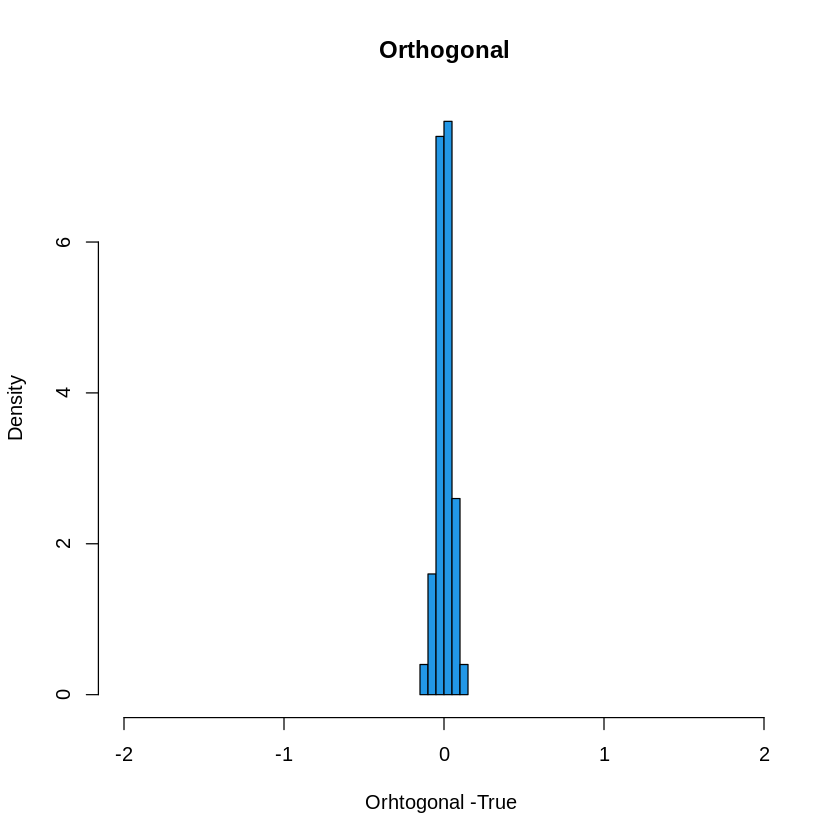
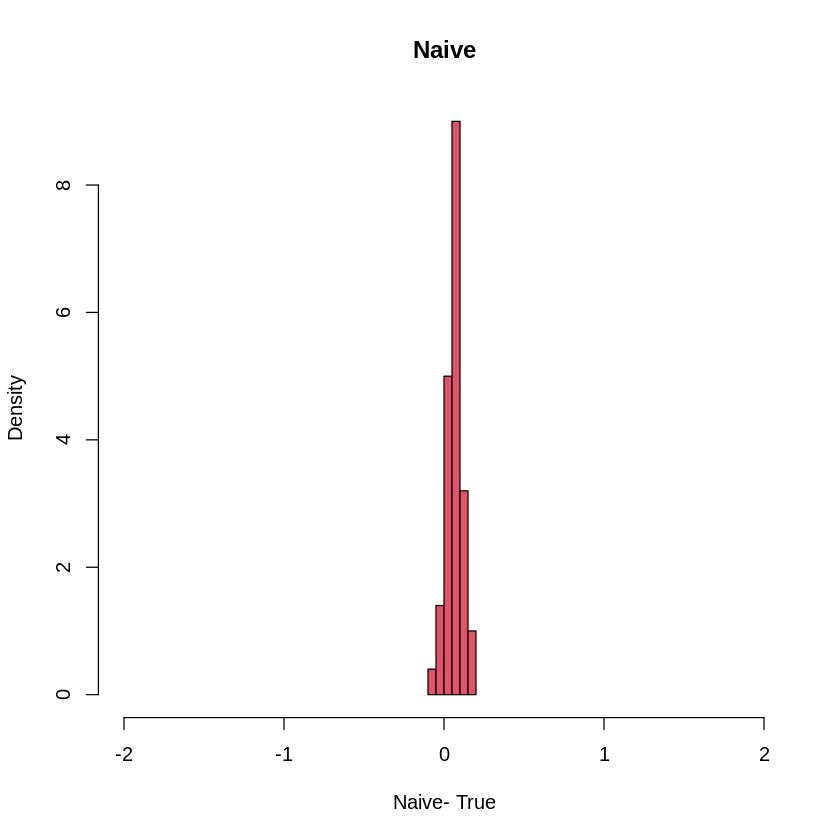
hist(Orthogonal-1,col=4, freq=F, xlim= c(-2, 2), xlab= "Orhtogonal -True ", main="Orthogonal")
hist(Naive-1, col=2, freq=F, xlim= c(-2,2), xlab= "Naive- True", main = "Naive")