25. Weak IV Experiments#
Simulation Design
# Simulation Design
install.packages("librarian")
librarian::shelf(hdm)
set.seed(1)
B= 10000 # trials
IVEst = rep(0, B)
n=100
beta = .25 # .2 weak IV
#beta = 1 # 1 strong IV
U = rnorm(n)
Z = rnorm(n) #generate instrument
D = beta*Z + U #generate endogenougs variable
Y = D+ U # the true causal effect is 1
summary(lm(D~Z)) # first stage is very weak here
summary(tsls(x=NULL, d=D, y=Y, z=Z)) #
Installing package into ‘/usr/local/lib/R/site-library’
(as ‘lib’ is unspecified)
also installing the dependency ‘BiocManager’
These packages will be installed:
'hdm'
It may take some time.
also installing the dependencies ‘iterators’, ‘foreach’, ‘shape’, ‘Rcpp’, ‘RcppEigen’, ‘glmnet’, ‘checkmate’, ‘Formula’
Call:
lm(formula = D ~ Z)
Residuals:
Min 1Q Median 3Q Max
-2.32416 -0.60361 0.00536 0.58305 2.29316
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 0.10885 0.09035 1.205 0.23118
Z 0.24907 0.09472 2.629 0.00993 **
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Residual standard error: 0.9028 on 98 degrees of freedom
Multiple R-squared: 0.0659, Adjusted R-squared: 0.05637
F-statistic: 6.914 on 1 and 98 DF, p-value: 0.009931
[1] "Estimates and Significance Testing from from tsls"
Estimate Std. Error t value p value
d1 0.99626 0.38173 2.610 0.00906 **
(Intercept) 0.10926 0.09824 1.112 0.26608
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Note that the instrument is weak here (contolled by \(\beta\)) – the t-stat is less than 4.
25.1. Run 1000 trials to evaluate distribution of the IV estimator#
# Simulation Design
set.seed(1)
B= 10000 # trials
IVEst = rep(0, B)
for(i in 1:B){
U = rnorm(n)
Z = rnorm(n) #generate instrument
D = beta*Z + U #generate endogenougs variable
Y = D+ U # the true causal effect is 1
IVEst[i] = coef(tsls(x=NULL, d=D, y=Y, z=Z))[1,1]
}
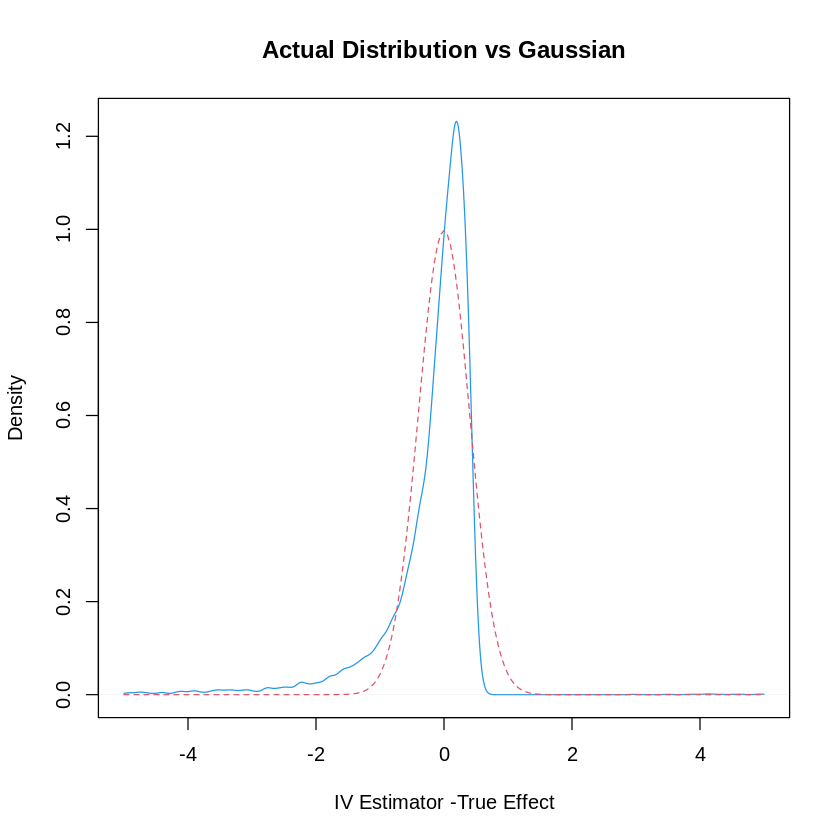
25.2. Plot the Actual Distribution against the Normal Approximation (based on Strong Instrument Assumption)#
plot(density(IVEst-1, n=1000, from=-5, to=5),col=4, xlim= c(-5, 5),
xlab= "IV Estimator -True Effect", main="Actual Distribution vs Gaussian")
val=seq(-5, 5, by=.05)
var = (1/beta^2)*(1/100) # theoretical variance of IV
sd = sqrt(var)
lines(val, dnorm(val, sd=sd), col=2, lty=2)
rejection.frequency = sum(( abs(IVEst-1)/sd > 1.96))/B
cat(c("Rejection Frequency is ", rejection.frequency, " while we expect it to be .05"))
Rejection Frequency is 0.1401 while we expect it to be .05
25.2.1. Some Help Functions#
# help(tsls)
# help(density)


